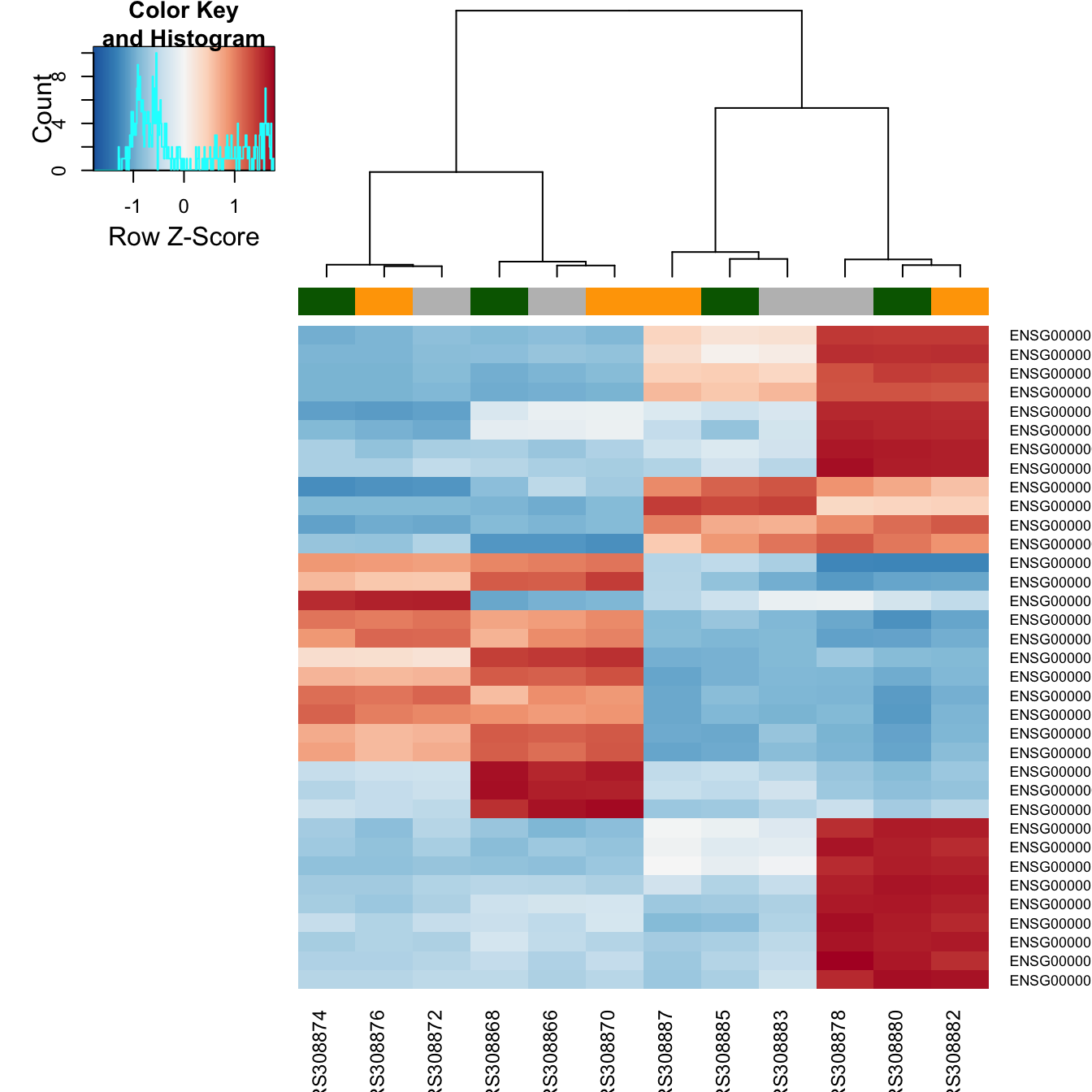
Differential Gene Expression Analysis Using Deseq2 . We will start from the fastq files, align to the reference genome,. This document presents an rnaseq differential expression workflow. Understanding the different steps in a differential expression. The standard differential expression analysis steps are wrapped into a single function, deseq. Its differential expression tests are based on a negative binomial generalized linear model. Deseq2 is a popular differential expression analysis package available through bioconductor.
from www.sthda.com
Understanding the different steps in a differential expression. The standard differential expression analysis steps are wrapped into a single function, deseq. This document presents an rnaseq differential expression workflow. Deseq2 is a popular differential expression analysis package available through bioconductor. Its differential expression tests are based on a negative binomial generalized linear model. We will start from the fastq files, align to the reference genome,.
RNASeq differential expression work flow using DESeq2 Easy Guides
Differential Gene Expression Analysis Using Deseq2 The standard differential expression analysis steps are wrapped into a single function, deseq. Understanding the different steps in a differential expression. We will start from the fastq files, align to the reference genome,. Deseq2 is a popular differential expression analysis package available through bioconductor. Its differential expression tests are based on a negative binomial generalized linear model. The standard differential expression analysis steps are wrapped into a single function, deseq. This document presents an rnaseq differential expression workflow.
From www.mdpi.com
Life Free FullText of scRNAseq Differential Gene Differential Gene Expression Analysis Using Deseq2 Understanding the different steps in a differential expression. This document presents an rnaseq differential expression workflow. The standard differential expression analysis steps are wrapped into a single function, deseq. We will start from the fastq files, align to the reference genome,. Its differential expression tests are based on a negative binomial generalized linear model. Deseq2 is a popular differential expression. Differential Gene Expression Analysis Using Deseq2.
From www.youtube.com
How I analyze RNA Seq Gene Expression data using DESeq2 YouTube Differential Gene Expression Analysis Using Deseq2 Its differential expression tests are based on a negative binomial generalized linear model. Understanding the different steps in a differential expression. This document presents an rnaseq differential expression workflow. The standard differential expression analysis steps are wrapped into a single function, deseq. Deseq2 is a popular differential expression analysis package available through bioconductor. We will start from the fastq files,. Differential Gene Expression Analysis Using Deseq2.
From www.researchgate.net
Differential expression analysis across multiple transcriptomewide Differential Gene Expression Analysis Using Deseq2 This document presents an rnaseq differential expression workflow. The standard differential expression analysis steps are wrapped into a single function, deseq. Understanding the different steps in a differential expression. We will start from the fastq files, align to the reference genome,. Its differential expression tests are based on a negative binomial generalized linear model. Deseq2 is a popular differential expression. Differential Gene Expression Analysis Using Deseq2.
From hbctraining.github.io
Genelevel differential expression analysis with DESeq2 Introduction Differential Gene Expression Analysis Using Deseq2 We will start from the fastq files, align to the reference genome,. Its differential expression tests are based on a negative binomial generalized linear model. Understanding the different steps in a differential expression. Deseq2 is a popular differential expression analysis package available through bioconductor. The standard differential expression analysis steps are wrapped into a single function, deseq. This document presents. Differential Gene Expression Analysis Using Deseq2.
From dmgatti.github.io
06 Differential expression analysis Introduction to RNAseq Differential Gene Expression Analysis Using Deseq2 The standard differential expression analysis steps are wrapped into a single function, deseq. Understanding the different steps in a differential expression. This document presents an rnaseq differential expression workflow. Deseq2 is a popular differential expression analysis package available through bioconductor. Its differential expression tests are based on a negative binomial generalized linear model. We will start from the fastq files,. Differential Gene Expression Analysis Using Deseq2.
From www.researchgate.net
RNAseq and DEG analysis. (A) Comparison of FLNB expression levels Differential Gene Expression Analysis Using Deseq2 Deseq2 is a popular differential expression analysis package available through bioconductor. The standard differential expression analysis steps are wrapped into a single function, deseq. Understanding the different steps in a differential expression. This document presents an rnaseq differential expression workflow. Its differential expression tests are based on a negative binomial generalized linear model. We will start from the fastq files,. Differential Gene Expression Analysis Using Deseq2.
From www.researchgate.net
Differential expression analysis data using DESeq2 Download Differential Gene Expression Analysis Using Deseq2 Deseq2 is a popular differential expression analysis package available through bioconductor. This document presents an rnaseq differential expression workflow. Its differential expression tests are based on a negative binomial generalized linear model. The standard differential expression analysis steps are wrapped into a single function, deseq. Understanding the different steps in a differential expression. We will start from the fastq files,. Differential Gene Expression Analysis Using Deseq2.
From www.researchgate.net
Differential expression analysis using DESeq2 in R. Download Differential Gene Expression Analysis Using Deseq2 Its differential expression tests are based on a negative binomial generalized linear model. We will start from the fastq files, align to the reference genome,. This document presents an rnaseq differential expression workflow. Understanding the different steps in a differential expression. The standard differential expression analysis steps are wrapped into a single function, deseq. Deseq2 is a popular differential expression. Differential Gene Expression Analysis Using Deseq2.
From www.researchgate.net
Analysis of differential gene expression profiles before and after the Differential Gene Expression Analysis Using Deseq2 Understanding the different steps in a differential expression. Its differential expression tests are based on a negative binomial generalized linear model. This document presents an rnaseq differential expression workflow. The standard differential expression analysis steps are wrapped into a single function, deseq. We will start from the fastq files, align to the reference genome,. Deseq2 is a popular differential expression. Differential Gene Expression Analysis Using Deseq2.
From www.researchgate.net
Differential gene expression analysis by RNAsequencing reveals Differential Gene Expression Analysis Using Deseq2 The standard differential expression analysis steps are wrapped into a single function, deseq. We will start from the fastq files, align to the reference genome,. Deseq2 is a popular differential expression analysis package available through bioconductor. This document presents an rnaseq differential expression workflow. Understanding the different steps in a differential expression. Its differential expression tests are based on a. Differential Gene Expression Analysis Using Deseq2.
From kyloot.com
RNASeq differential expression work flow using DESeq2 Easy Guides Differential Gene Expression Analysis Using Deseq2 This document presents an rnaseq differential expression workflow. Deseq2 is a popular differential expression analysis package available through bioconductor. The standard differential expression analysis steps are wrapped into a single function, deseq. Its differential expression tests are based on a negative binomial generalized linear model. Understanding the different steps in a differential expression. We will start from the fastq files,. Differential Gene Expression Analysis Using Deseq2.
From bioinformaticsworkbook.org
RNA Sequence Analysis Bioinformatics Workbook Differential Gene Expression Analysis Using Deseq2 Its differential expression tests are based on a negative binomial generalized linear model. We will start from the fastq files, align to the reference genome,. The standard differential expression analysis steps are wrapped into a single function, deseq. Understanding the different steps in a differential expression. Deseq2 is a popular differential expression analysis package available through bioconductor. This document presents. Differential Gene Expression Analysis Using Deseq2.
From biocorecrg.github.io
Differential gene expression Differential Gene Expression Analysis Using Deseq2 Its differential expression tests are based on a negative binomial generalized linear model. This document presents an rnaseq differential expression workflow. Deseq2 is a popular differential expression analysis package available through bioconductor. We will start from the fastq files, align to the reference genome,. Understanding the different steps in a differential expression. The standard differential expression analysis steps are wrapped. Differential Gene Expression Analysis Using Deseq2.
From www.researchgate.net
Differential gene expression analysis. ac Principal component analysis Differential Gene Expression Analysis Using Deseq2 The standard differential expression analysis steps are wrapped into a single function, deseq. This document presents an rnaseq differential expression workflow. Deseq2 is a popular differential expression analysis package available through bioconductor. We will start from the fastq files, align to the reference genome,. Its differential expression tests are based on a negative binomial generalized linear model. Understanding the different. Differential Gene Expression Analysis Using Deseq2.
From www.youtube.com
Differential Gene Expression Bulk RNA Seq Analysis Results (DESEQ2 Differential Gene Expression Analysis Using Deseq2 This document presents an rnaseq differential expression workflow. We will start from the fastq files, align to the reference genome,. The standard differential expression analysis steps are wrapped into a single function, deseq. Its differential expression tests are based on a negative binomial generalized linear model. Understanding the different steps in a differential expression. Deseq2 is a popular differential expression. Differential Gene Expression Analysis Using Deseq2.
From www.researchgate.net
Differential gene expression analysis. (A) Principal component analysis Differential Gene Expression Analysis Using Deseq2 The standard differential expression analysis steps are wrapped into a single function, deseq. We will start from the fastq files, align to the reference genome,. Its differential expression tests are based on a negative binomial generalized linear model. This document presents an rnaseq differential expression workflow. Deseq2 is a popular differential expression analysis package available through bioconductor. Understanding the different. Differential Gene Expression Analysis Using Deseq2.
From www.researchgate.net
Differential gene expression analysis (DESeq2) shows significant gene Differential Gene Expression Analysis Using Deseq2 Deseq2 is a popular differential expression analysis package available through bioconductor. We will start from the fastq files, align to the reference genome,. This document presents an rnaseq differential expression workflow. Its differential expression tests are based on a negative binomial generalized linear model. Understanding the different steps in a differential expression. The standard differential expression analysis steps are wrapped. Differential Gene Expression Analysis Using Deseq2.
From www.researchgate.net
Differential gene expression between adult and aging microglia. ad Differential Gene Expression Analysis Using Deseq2 This document presents an rnaseq differential expression workflow. Its differential expression tests are based on a negative binomial generalized linear model. Understanding the different steps in a differential expression. We will start from the fastq files, align to the reference genome,. The standard differential expression analysis steps are wrapped into a single function, deseq. Deseq2 is a popular differential expression. Differential Gene Expression Analysis Using Deseq2.